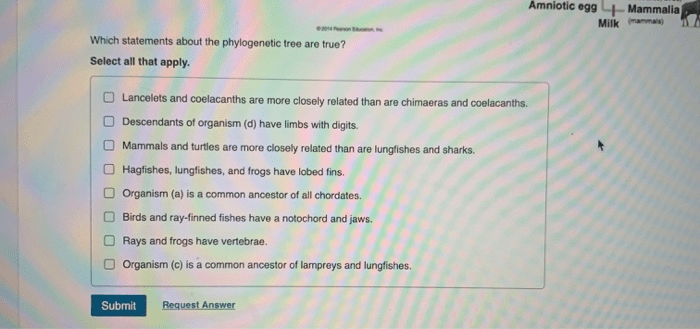
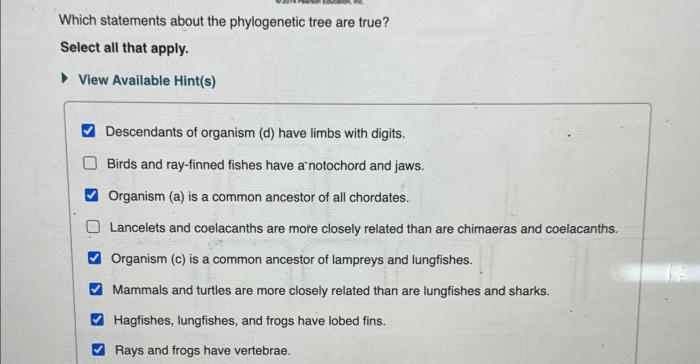
Which statements about the phylogenetic tree are true? This article delves into the fascinating world of phylogenetic trees, exploring their construction, interpretation, and applications in evolutionary biology. From understanding the hierarchical structure of these trees to interpreting branch lengths and identifying common ancestors, we will uncover the intricacies of phylogenetic trees and their significance in unraveling evolutionary history.
Phylogenetic trees are essential tools in evolutionary biology, providing valuable insights into the relationships between species and their evolutionary pathways. This comprehensive guide will equip you with a thorough understanding of phylogenetic trees, enabling you to navigate their complexities and appreciate their significance in scientific research.
Phylogenetic Tree Overview: Which Statements About The Phylogenetic Tree Are True
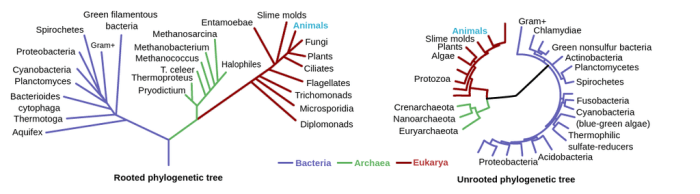
A phylogenetic tree is a diagram that represents the evolutionary relationships between different species or other taxa. It is a branching diagram that shows the hypothetical relationships between groups of organisms based on shared characteristics.
Phylogenetic trees are used in evolutionary biology to infer the evolutionary history of species and to understand the patterns and processes of evolution. They can be used to study a wide range of topics, including the origins of life, the evolution of new species, and the relationships between different groups of organisms.
A basic phylogenetic tree typically consists of a series of nodes connected by branches. The nodes represent the ancestors of the groups of organisms, and the branches represent the evolutionary relationships between the groups. The length of the branches can be used to infer the amount of evolutionary change that has occurred between the groups.
Key Characteristics of Phylogenetic Trees, Which statements about the phylogenetic tree are true
Phylogenetic trees have a hierarchical structure, with the root of the tree representing the most recent common ancestor of all the taxa in the tree. The nodes in the tree represent the ancestors of the groups of organisms, and the branches represent the evolutionary relationships between the groups.
The length of the branches in a phylogenetic tree can be used to infer the amount of evolutionary change that has occurred between the groups. The longer the branch, the more evolutionary change has occurred.
Phylogenetic trees can be used to identify common ancestors and descendant groups. The common ancestor of two groups is the most recent ancestor that they share. The descendant groups of an ancestor are all the groups that are descended from that ancestor.
Methods of Phylogenetic Tree Construction
There are a number of different methods for constructing phylogenetic trees. Some of the most common methods include maximum parsimony, neighbor-joining, and Bayesian inference.
Maximum parsimony is a method that assumes that the simplest explanation for the data is the most likely to be true. This method finds the tree that requires the fewest evolutionary changes to explain the data.
Neighbor-joining is a method that uses a distance matrix to construct a phylogenetic tree. The distance matrix contains the distances between all pairs of taxa in the tree.
Bayesian inference is a method that uses statistical methods to estimate the probability of different phylogenetic trees. This method takes into account the uncertainty in the data and the different possible evolutionary models.
Interpretation of Phylogenetic Trees
Phylogenetic trees can be used to infer evolutionary relationships between different species or other taxa. The branch lengths in a phylogenetic tree can be used to infer the amount of evolutionary change that has occurred between the groups.
Phylogenetic trees can also be used to identify common ancestors and descendant groups. The common ancestor of two groups is the most recent ancestor that they share. The descendant groups of an ancestor are all the groups that are descended from that ancestor.
Phylogenetic trees can be used to study evolutionary history and diversity. They can be used to understand the origins of life, the evolution of new species, and the relationships between different groups of organisms.
Limitations of Phylogenetic Trees
Phylogenetic trees are a powerful tool for studying evolutionary relationships, but they also have some limitations. One limitation is that they are based on incomplete data.
Another limitation of phylogenetic trees is that they are based on assumptions about the evolutionary process. These assumptions may not always be true, and this can lead to errors in the tree.
Despite these limitations, phylogenetic trees are a valuable tool for studying evolutionary relationships. They can provide insights into the origins of life, the evolution of new species, and the relationships between different groups of organisms.
Applications of Phylogenetic Trees
Phylogenetic trees have a wide range of applications in different fields of biology. They are used in taxonomy to classify organisms into different groups. They are also used in conservation biology to identify endangered species and to develop conservation strategies.
Phylogenetic trees are also used in medicine to study the evolution of diseases and to develop new treatments. They are also used in agriculture to study the evolution of crops and to develop new varieties.
Phylogenetic trees are a powerful tool for studying evolutionary relationships and they have a wide range of applications in different fields of biology.
FAQ Resource
What is a phylogenetic tree?
A phylogenetic tree is a diagram that represents the evolutionary relationships among different species or other taxa. It is a branching diagram that shows the inferred evolutionary history of a group of organisms, based on shared characteristics and genetic similarities.
How are phylogenetic trees constructed?
Phylogenetic trees can be constructed using various methods, such as maximum parsimony, neighbor-joining, and Bayesian inference. These methods analyze genetic data or other characteristics to infer the most likely evolutionary relationships among the taxa being studied.
What are the limitations of phylogenetic trees?
Phylogenetic trees are limited by the availability and quality of data, as well as the assumptions and algorithms used in their construction. They may not always accurately represent the true evolutionary history due to factors such as incomplete fossil records, hybridization, and genetic recombination.